Seriatopora spp.-rich reef
I froze samples from the Seriatopora hystrix variable temperature study (SHVTS) for years (nine to be exact), just hoping that proteomic methodologies would 1) improve and 2) drop in price. Thankfully, both happened, so I used funds from the Friendly Bear Editorial Service and crowd sourcing (thanks everyone!) to analyze the proteins synthesized by a subset of corals from the experiments: upwelling reef corals exposed to stable or variable temperatures and non-upwelling corals exposed to these same treatments. The manuscript was published in August 2020 in Microorganisms, and I have submitted all data (RAW MS files, MZID files, MZML files, and the corresponding tab-delineated spreadsheets) to three repositories: NOAA’s National Centers for Environmental Information, the University of California San Diego’s MASSive Repository (accession MSV000085863), and Proteome Xchange (accession: PXD020679).
Basically, of the thousands of proteins sequenced, I focused on 30; check on the breakdown in this figure. DCP=differentially concentrated proteins (i.e., the ones that responded to experimental treatment). POIs= “proteins of interest” (in a nutshell, the ones that are useful for predicting coral responses to environmental change).
Looking at the proteins in a multivariate context: principal components analysis (PCA) and multi-dimensional scaling (MDS; similarity). S=stable temperature samples. V=variable temperature samples. You can see some pretty good separation by site: Houbihu (upwelling reef; green) vs. Houwan (non-upwelling; black). The corals exposed to their “native” conditions are most distinct from one another: Houbihu-variable (green V’s) vs. Houwan-stable (black S’s).
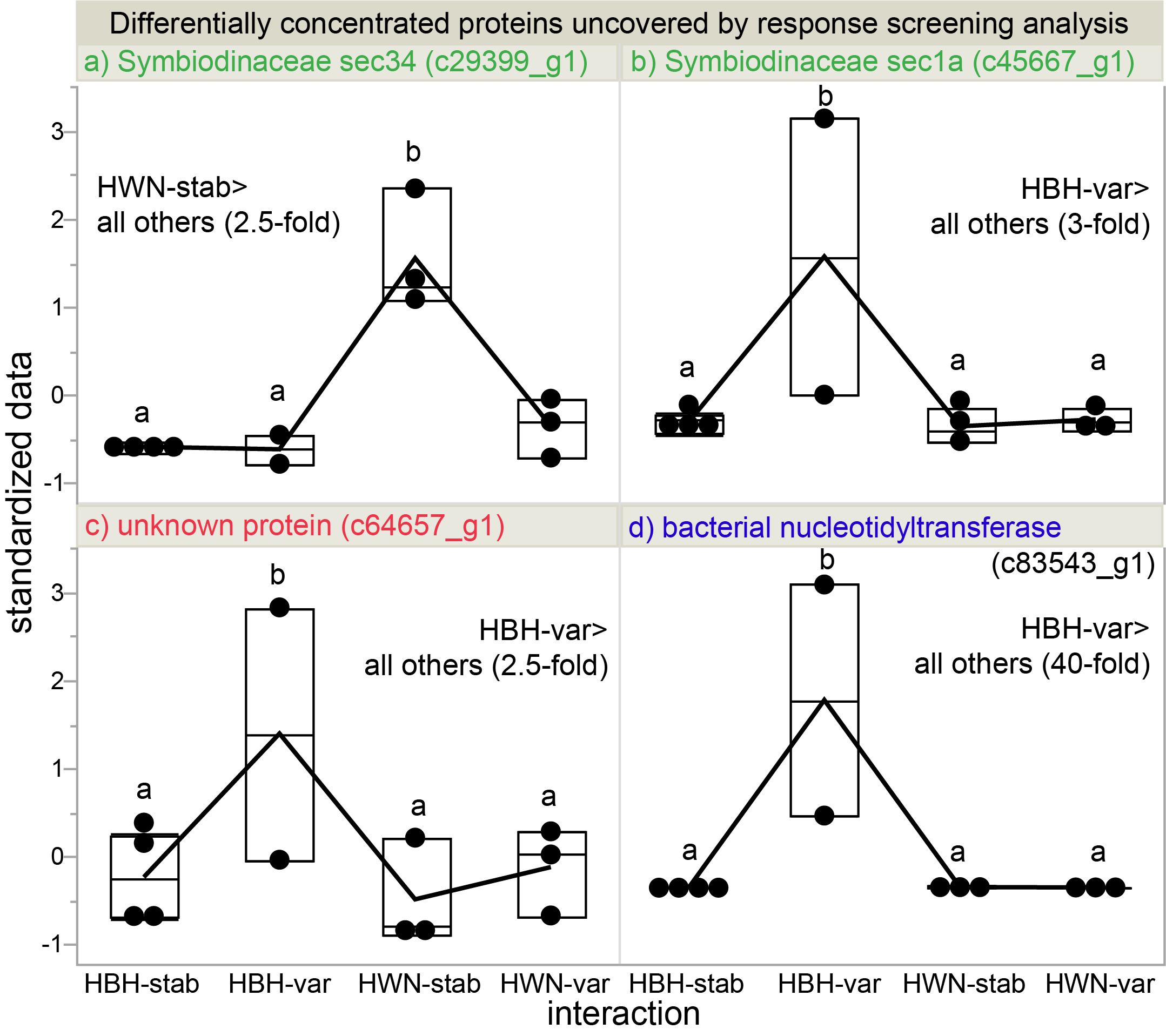
These are the proteins that essentially changed the most. I am particularly interested in the top two Symbiodinaceae proteins since they were both higher under native conditions and involved in the same cellular process: Golgi-mediated lipid and protein trafficking. We actually have a number of coral lipid trafficking papers, so we might be able to run with this (i.e., the role of this cellular process in thermal acclimation).
The final plots/data below are getting at one of my current (mid-2020) pet projects: predicting coral health. This particular technique, which is not the only one out there for this kind of exercise, is known as stepwise discriminant analysis (SDA). Basically, it is showing me the minimum number of proteins that can be used to build a statistical model that would permit me to predict the coral response to an experimental factor with 100% confidence. For this study, it may make less sense since it was carried out in a controlled laboratory setting and the data are inherently explanatory in nature. However, if you sampled a coral from a reef and did not know much about its thermal history, you could look at three proteins (the three vectors in b) and see if it was being exposed to stable or variable temperatures in the days leading up to sampling. Ultimately, this approach is more geared towards stress studies (i.e., estimating risk for corals exposed to, for instance, high temperatures for a prolonged period).