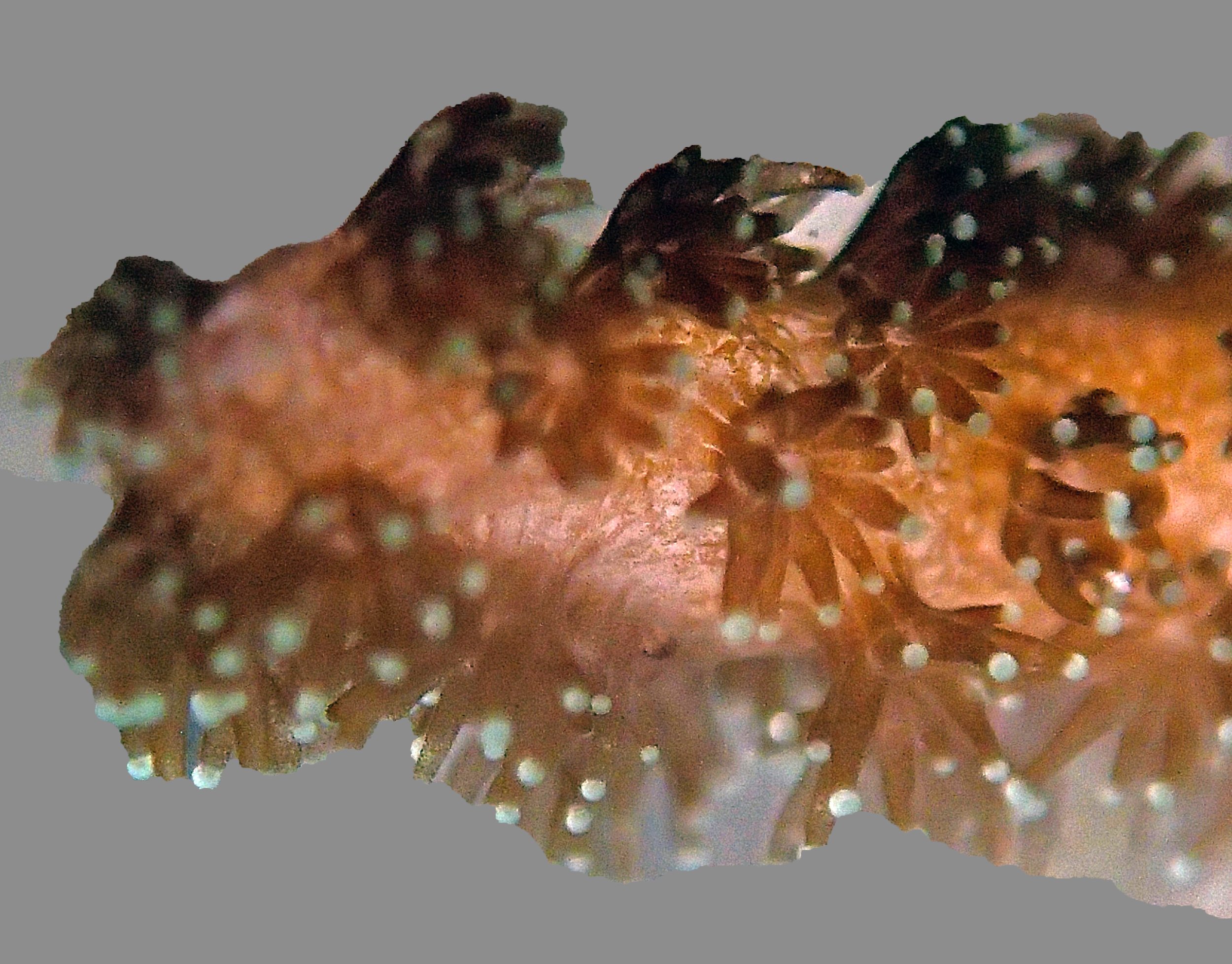
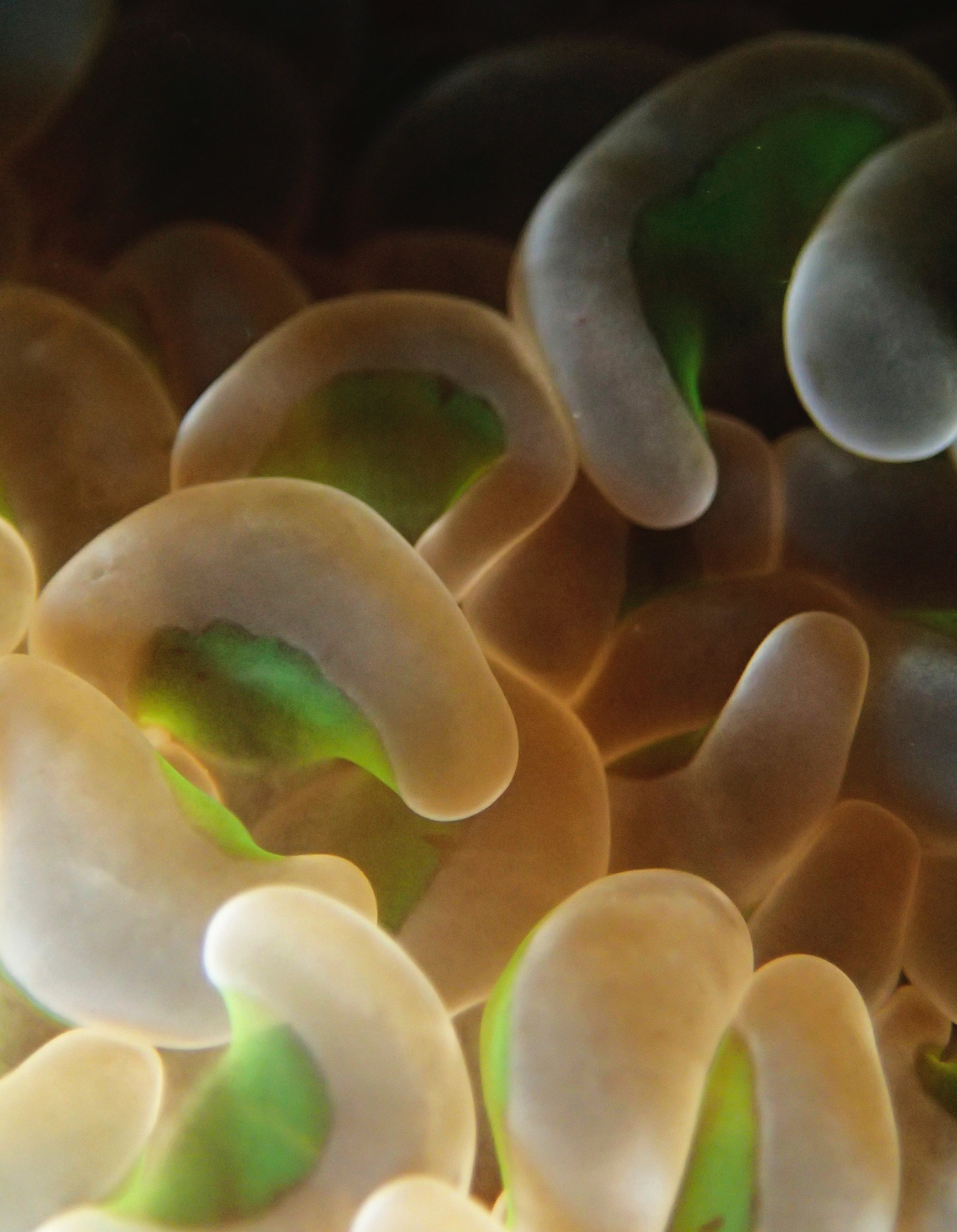
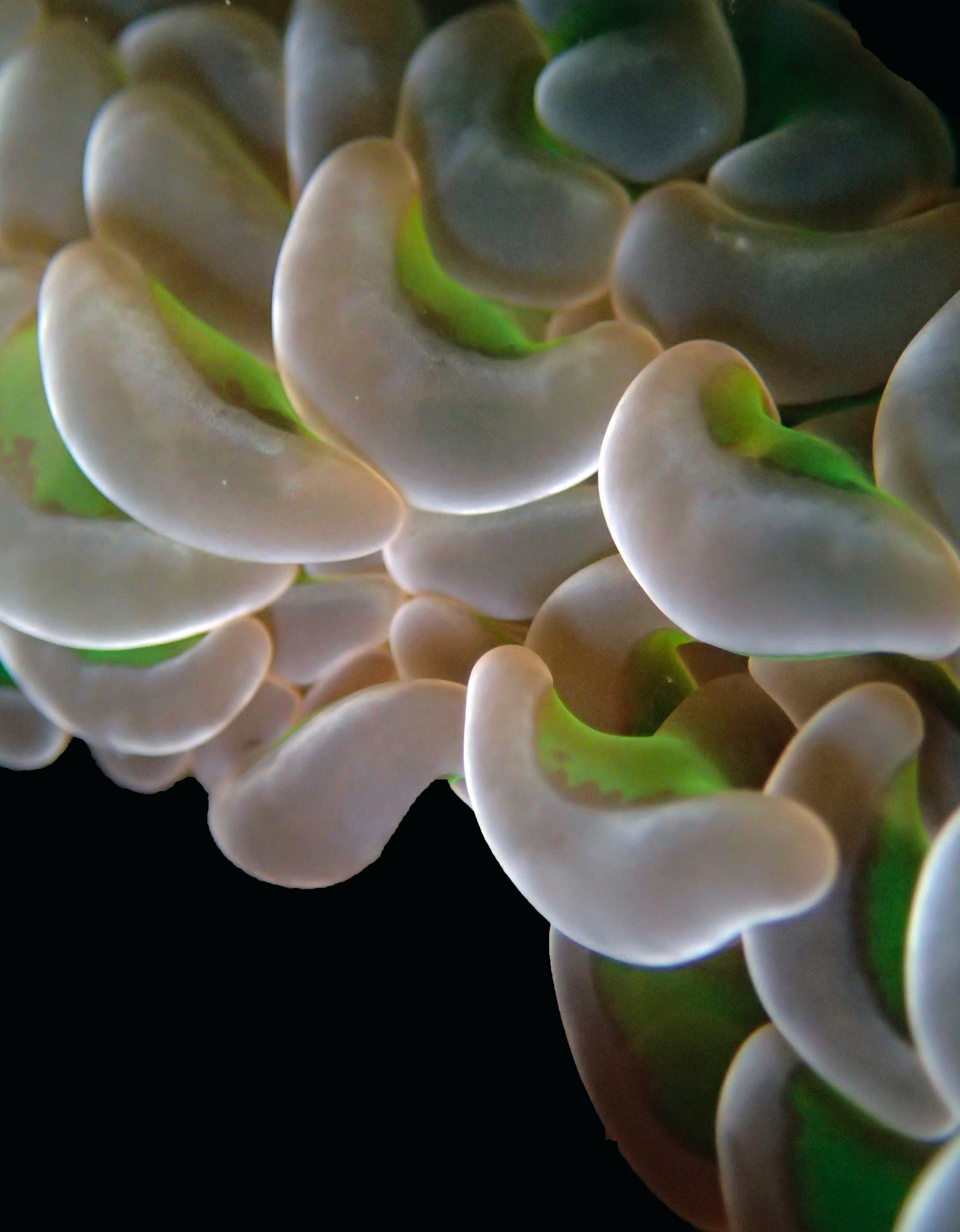
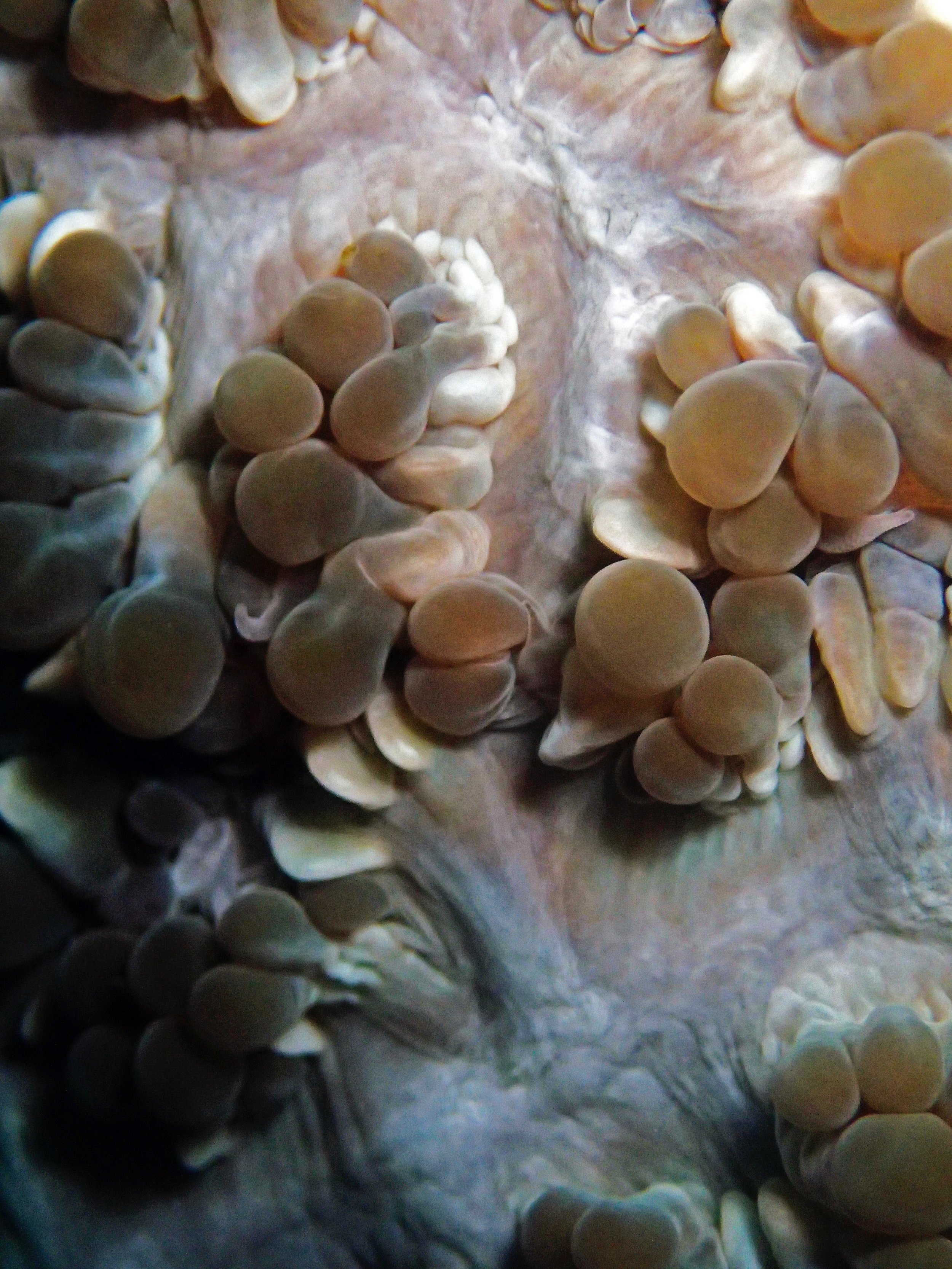
One day early in the trip (2015-1-10) and two later on (2015-2-8 to 2025-2-9) were spent surveying and sampling the bizarre, high-pCO2 reefs of stagnant, isolated, and hidden Nikko Bay. No Pocillopora damicornis colonies were sampled; all were later genotyped as Pocillopora acuta (using the Johnston et al. 2018 Peer J RFLP assay courtesy of Dr. Ben Wainwright). Since RFLPs were used for genotyping, rather than PCR+sequencing as for prior research cruises, no links to NCBI pages have been provided.

High-resolution habitat maps will be eventually placed on the Living Oceans Foundation’s mapping site.
PaNi01 (2015-1-10) lagoonal fringing reef
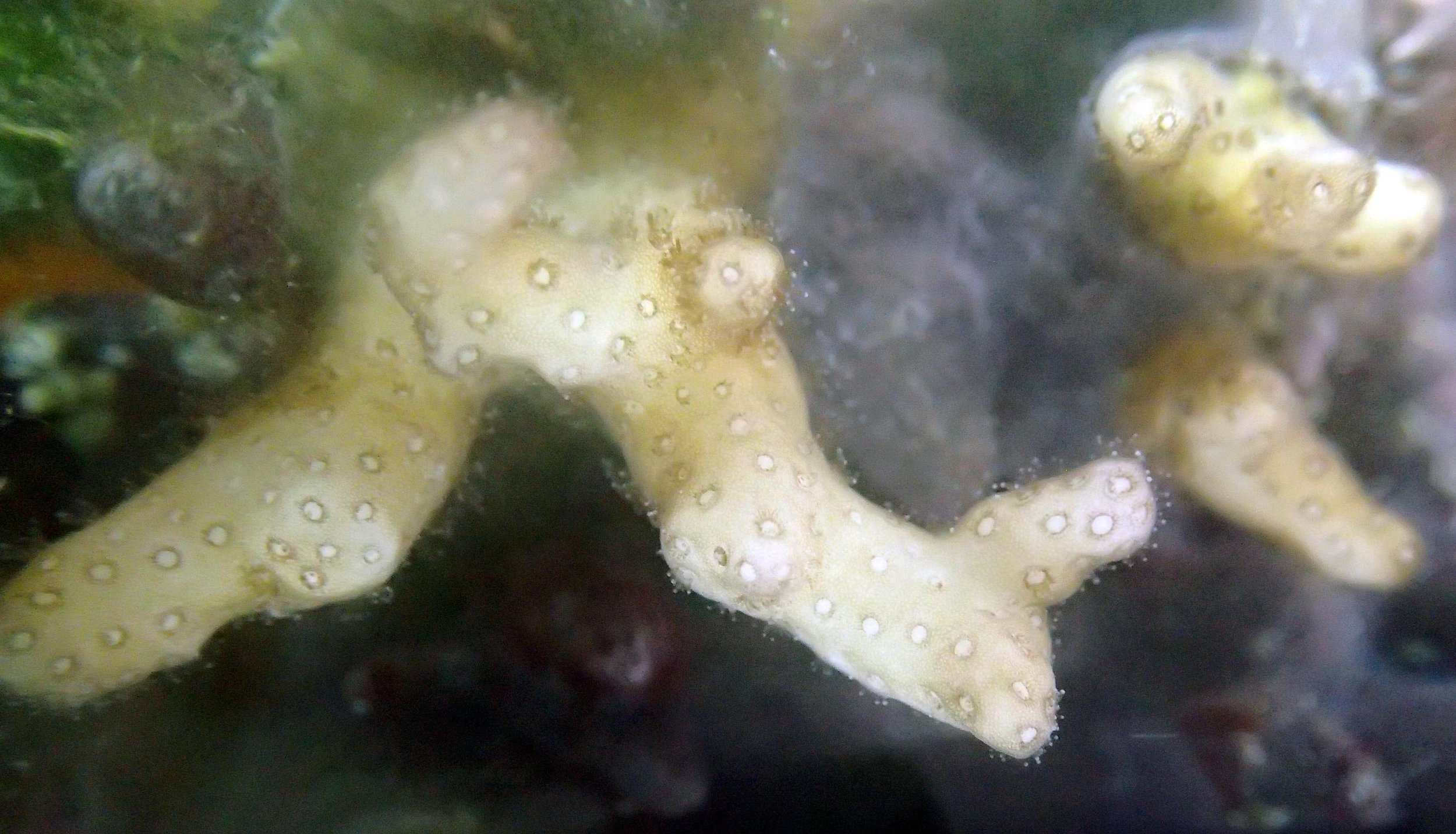
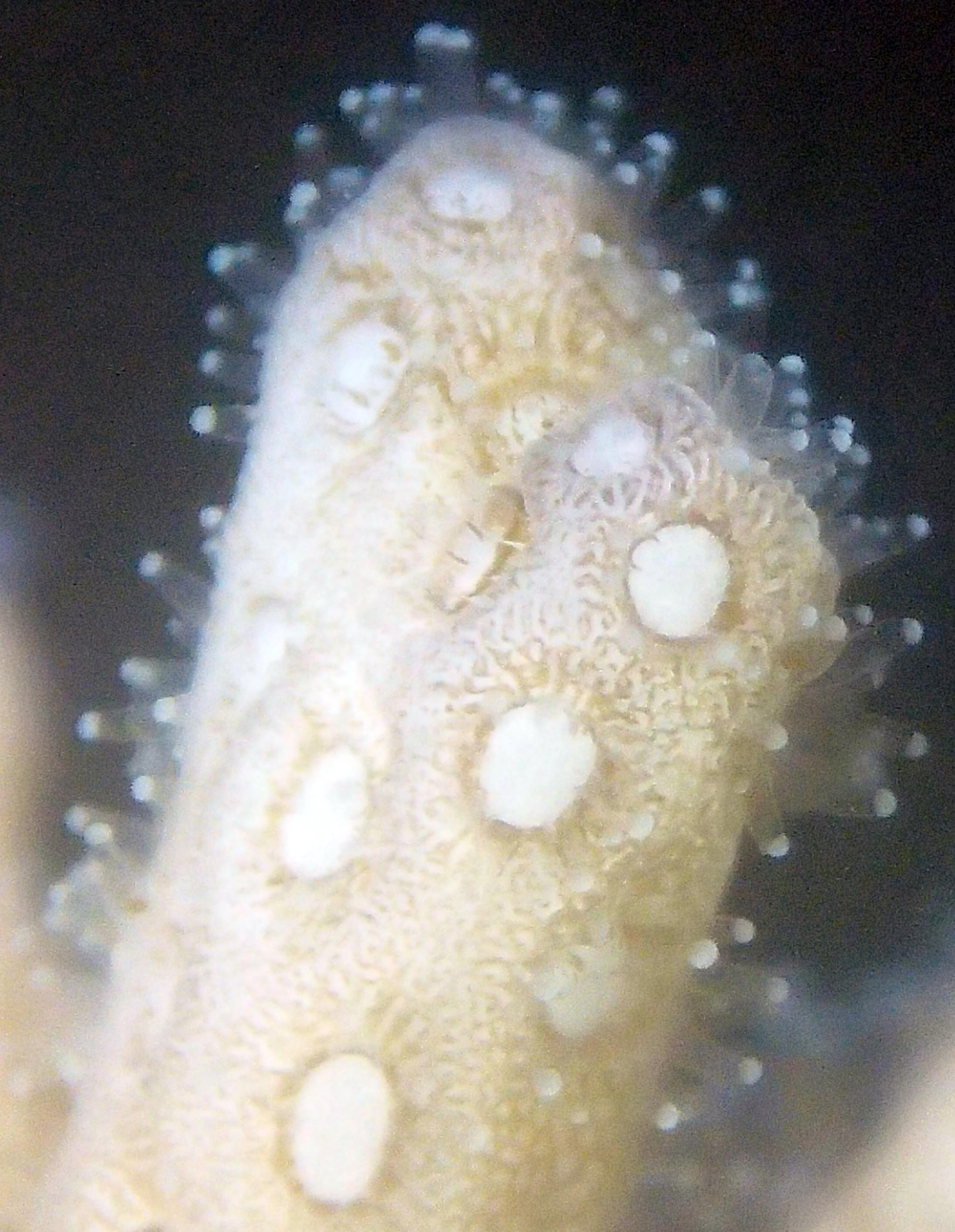
PaNi01 sampled pocilloporid colonies (n=3). For data from all colonies in the same file, please see this file.
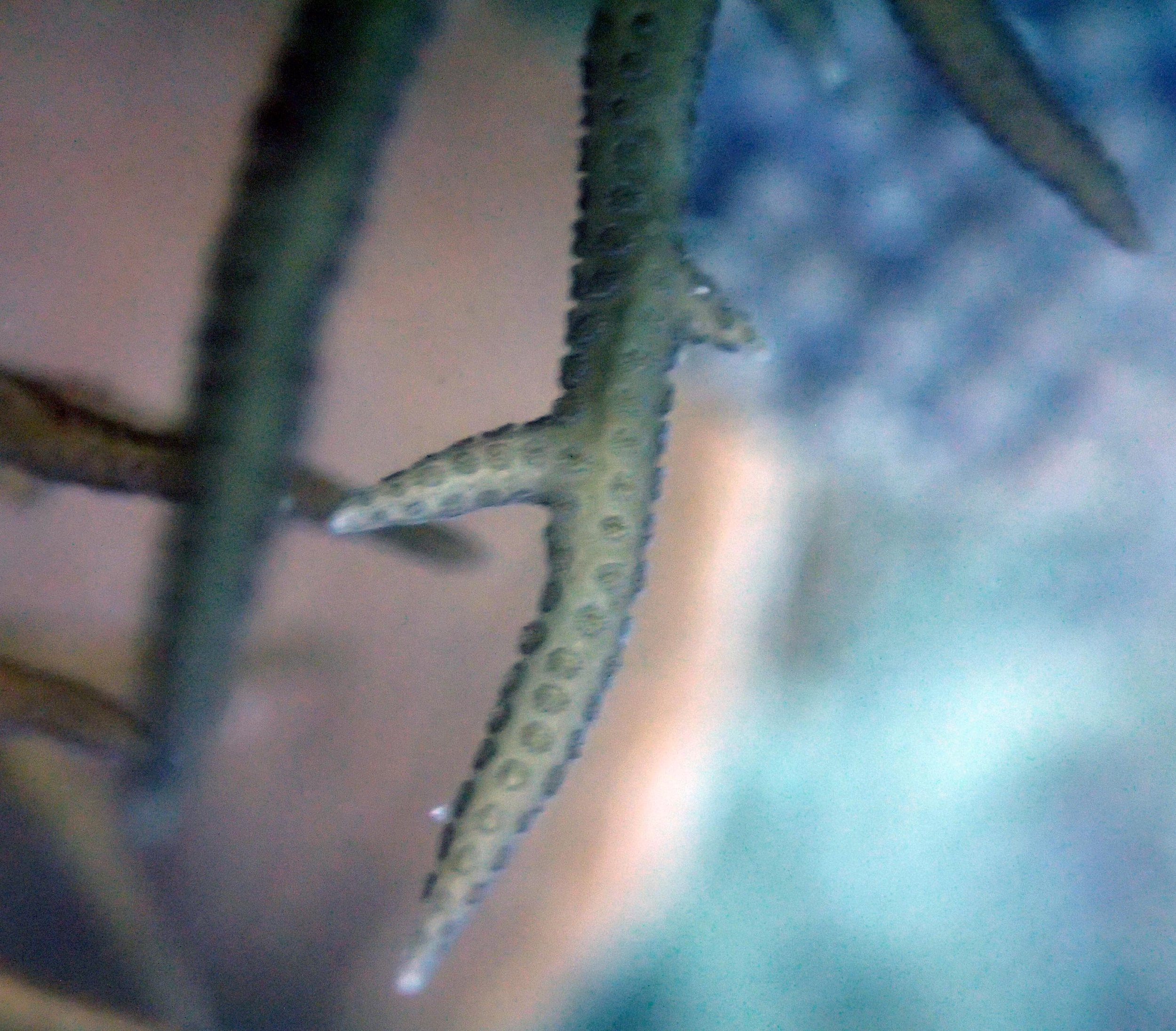
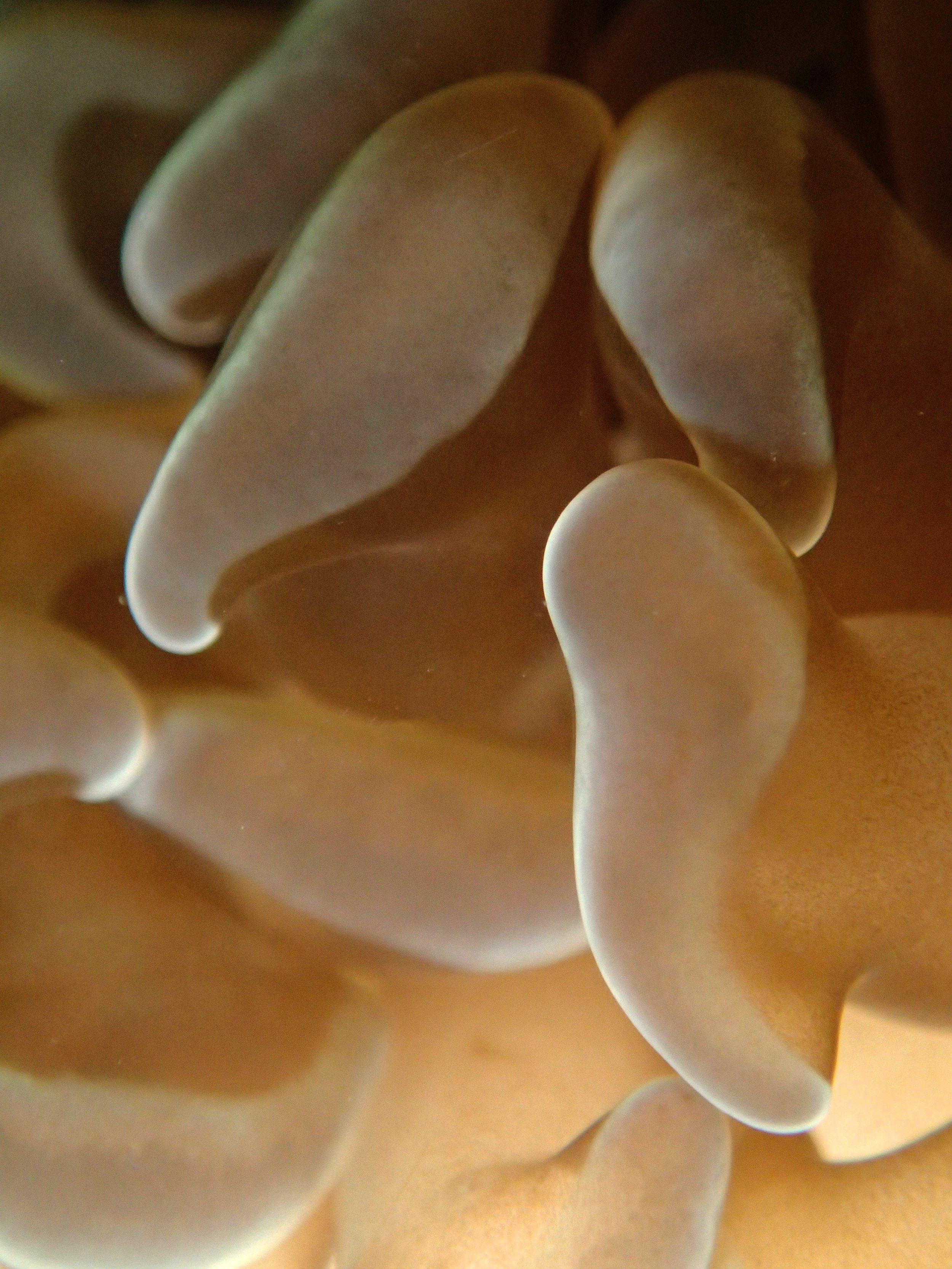
PaNi01 sampled Seriatopora hystrix colony (n=1)
Our return visit at the end of the mission (2015-2-8 to 2-9): PaNi85 (no pocilloporids sampled & no seriatoporids observed; note that this was virtually the same site as PaNi01.)
PaNi86 (2015-2-9): three pocilloporids were sampled (see below.) though no seriatoporids were observed at this lagoonal fringing reef.
PaNi86 sampled P. acuta colonies (n=3). Please note that, in general, I maintained the “Pd” sample code since, at that time, we were not yet confident in our ability to distinguish P. damicornis from P. acuta.

Click here for data, though note that the biopsy from this colony has not yet been analyzed.





Click here for data, though note that the biopsy from this colony has not yet been analyzed.


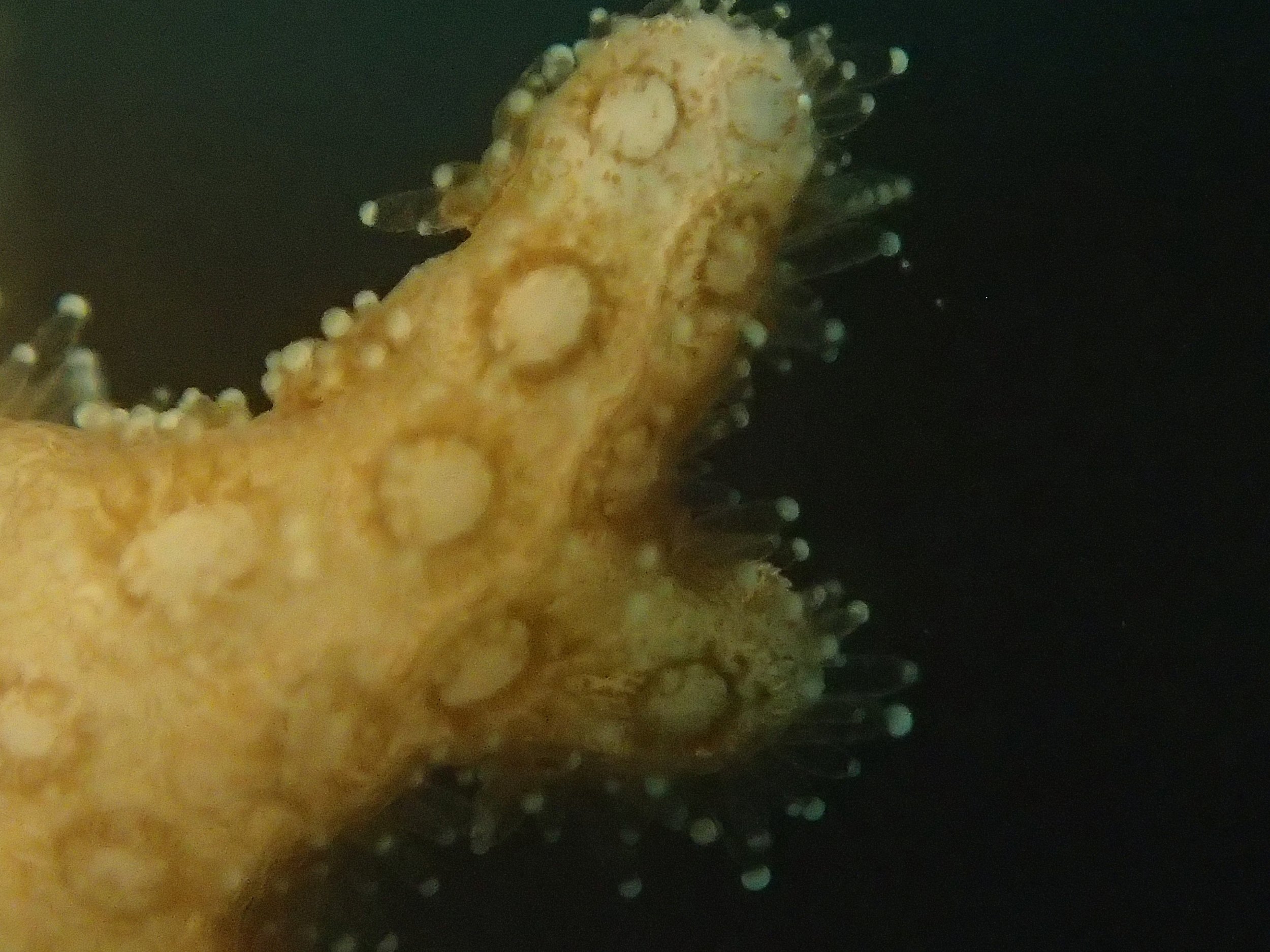
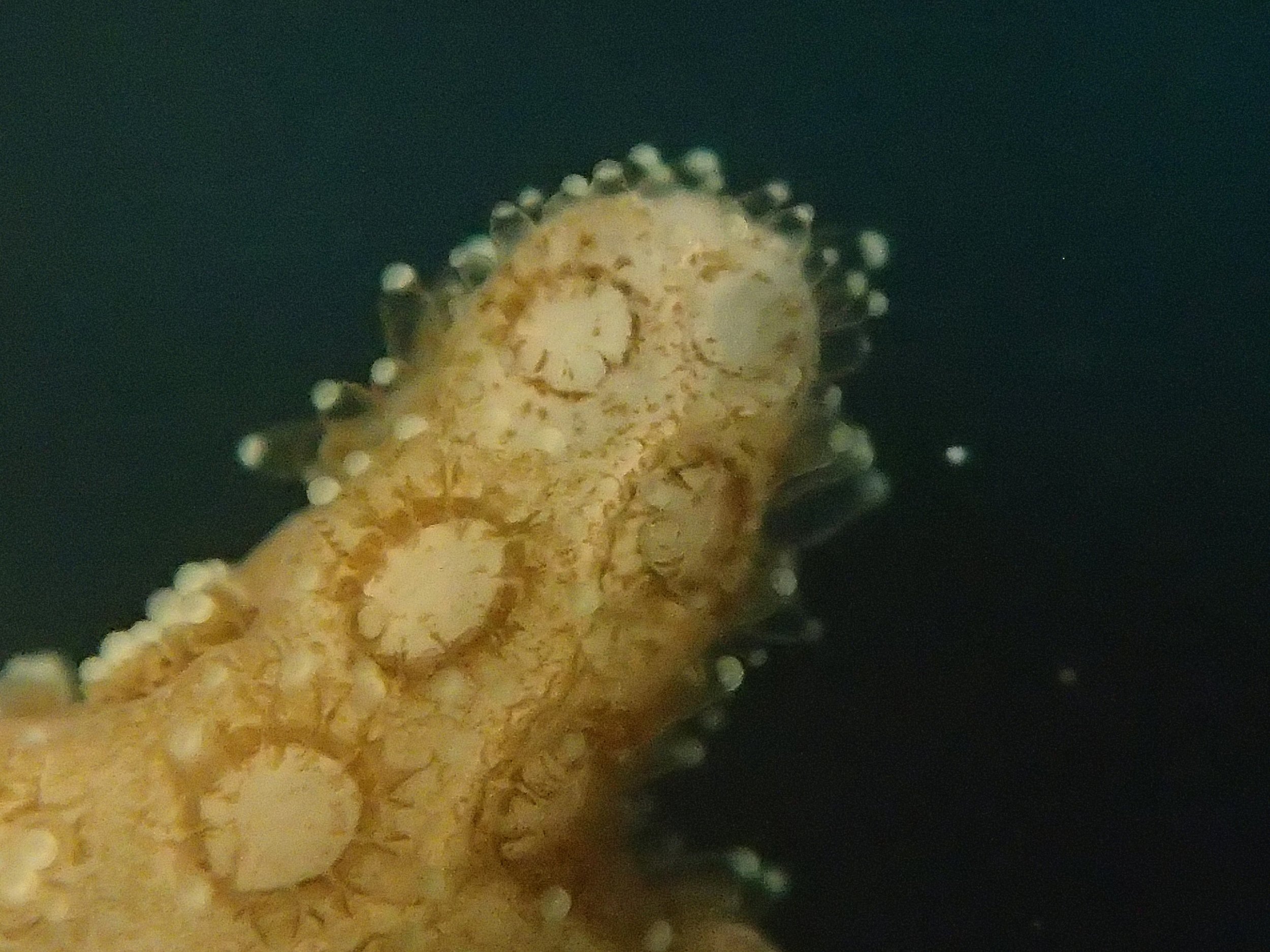
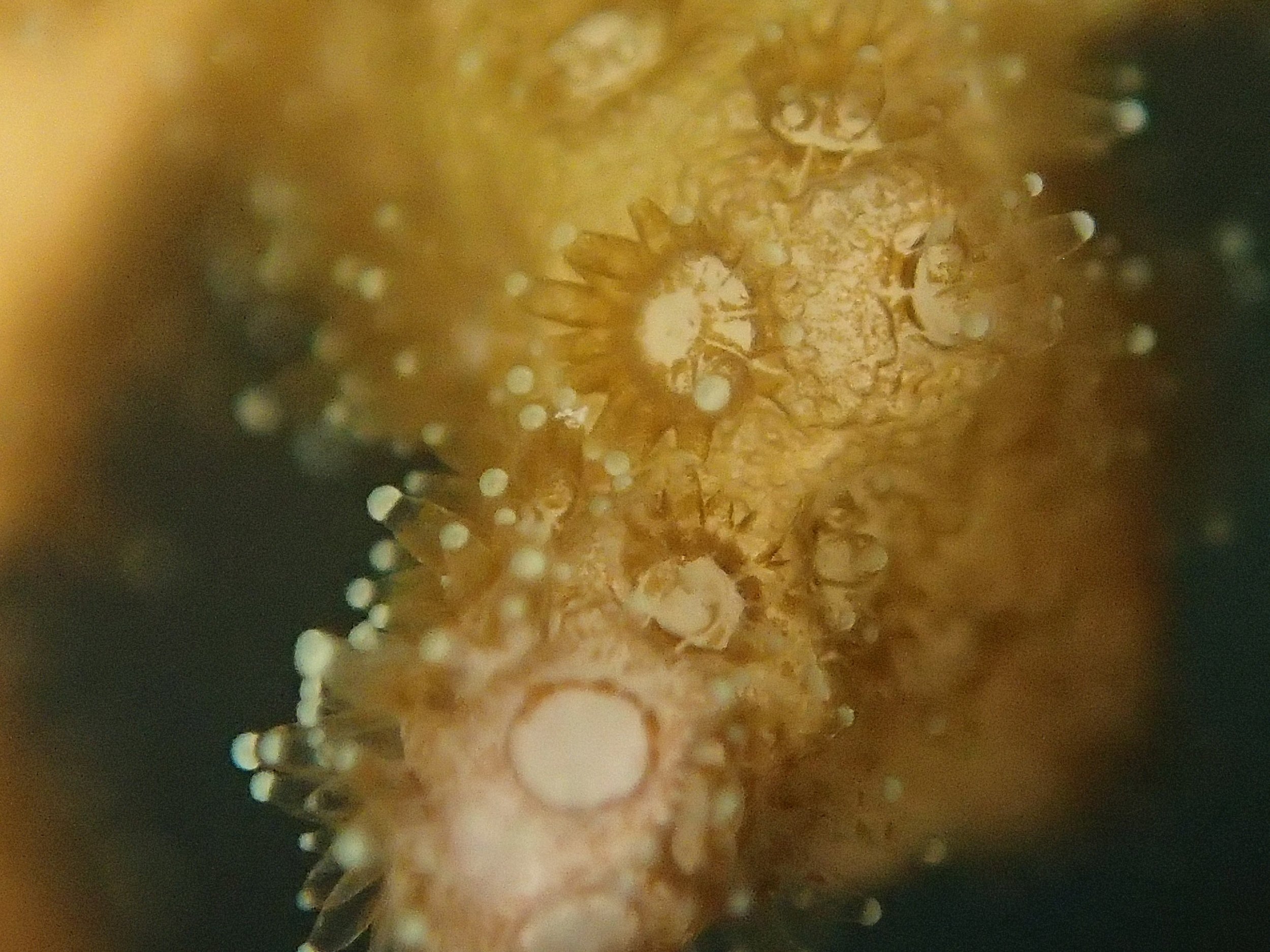

Click here for meta-data, though note that the biopsy from this colony has not yet been analyzed in full.

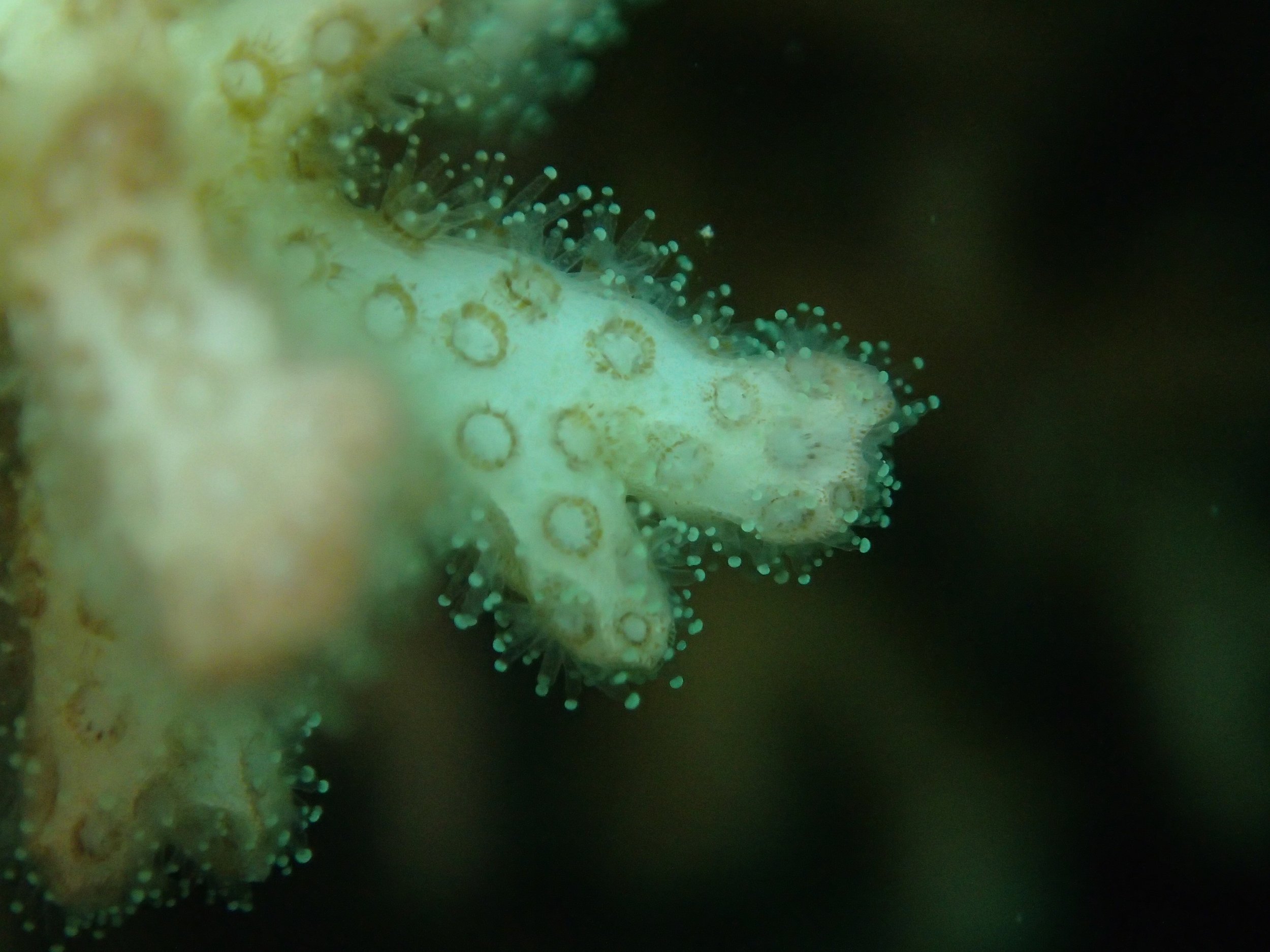
















PaNi87 (2015-2-9) P. acuta was common here but not sampled. Instead, I mainly sampled seriatoporid colonies (see images posted below the site pictures.) at this lagoonal fringing reef.
Seven seriatoporid colonies were sampled from PaNi87 (PaSe211-217), though images were not taken of the final one (PaSe217; click here for its respective meta-data).